Abstract
Tissue engineering is a broad field with applications ranging from pharmaceutical testing to total organ replacement. Recently, there has been extensive research on creating tissue that is able to replace or repair natural human tissue. Much of this research focuses on the creation of scaffolds that can both support cell growth and successfully integrate with the surrounding tissue. This article will introduce the concept of a scaffold for tissue engineering; discuss key areas of research including biomolecule use, vascularization, mechanical strength, and tissue attachment; and introduce some important recent advancements in these areas.
Introduction
Tissue engineering relies on four main factors: the growth of appropriate cells, the introduction of the proper biomolecules to these cells, the attachment of the cells to an appropriate scaffold, and the application of specific mechanical and biological forces to develop the completed tissue.1
Successful cell culture has been possible since the 1960’s, but these early methods lacked the adaptability necessary to make functioning tissues. With the introduction of induced pluripotent stem cells in 2008, however, researchers have not faced the same resource limitation previously encountered. As a result, the growth of cells of a desired type has not been limiting to researchers in tissue engineering and thus warrants less concern than other factors in contemporary tissue engineering.2,3
Similarly, the introduction of essential biomolecules (such as growth factors) to the developing tissue has generally not restricted modern tissue engineering efforts. Extensive research and knowledge of biomolecule function as well as relatively reliable methods of obtaining important biomolecules have allowed researchers to make engineered tissues more successfully emulate functional human tissue using biomolecules.4,5 Despite these advancements in information and procurement methods, however, the ability of biomolecules to improve engineered tissue often relies on the structure and chemical composition of the scaffold material.6
Cellular attachment has also been a heavily explored field of research. This refers specifically to the ability of the engineered tissue to seamlessly integrate into the surrounding tissue. Studies in cellular attachment often focus on qualities of scaffolds such as porosity as well as the introduction of biomolecules to encourage tissue union on the cellular level. Like biomolecule effectiveness, successful cellular attachment depends on the material and structure of the tissue scaffolding.7
Also critical to developing functional tissue is exposing it to the right environment. This development of tissue properties via the application of mechanical and biological forces depends strongly on finding materials that can withstand the required forces while supplying cells with the necessary environment and nutrients. Previous research in this has focused on several scaffold materials for various reasons. However, improvements to the material or the specific methods of development are still greatly needed in order to create functional implantable tissue. Because of the difficulty of conducting research in this area, devoted efforts to improving these methods remain critical to successful tissue engineering.
In order for a scaffold to be capable of supporting cells until the formation of a functioning tissue, it is necessary to satisfy several key requirements, principally introduction of helpful biomolecules, vascularization, mechanical function, appropriate chemical and physical environment, and compatibility with surrounding biological tissue.8,9 Great progress has been made towards satisfying many of these conditions, but further research in the field of tissue engineering must address challenges with existing scaffolds and improve their utility for replacing or repairing human tissue.
Key Research Areas of Scaffolding Design
Biomolecules
Throughout most early tissue engineering projects, researchers focused on simple cell culture surrounding specific material scaffolds.10 Promising developments such as the creation of engineered cartilage motivated further funding and interest in research. However, these early efforts missed out on several crucial factors to tissue engineering that allow implantable tissue to take on more complex functional roles. In order to create tissue that is functional and able to direct biological processes alongside nearby natural tissue, it is important to understand the interactions of biomolecules with engineered tissue.
Because the ultimate goal of tissue engineering is to create functional, implantable tissue that mimics biological systems, most important biomolecules have been explored by researchers in the medical field outside of tissue engineering. As a result, a solid body of research exists describing the functions and interactions of various biomolecules. Because of this existing information, understanding their potential uses in tissue engineering relies mainly on studying the interactions of biomolecules with materials which are not native to the body; most commonly, these non-biological materials are used as scaffolding. To complicate the topic further, biomolecules are a considerably large category encompassing everything from DNA to glucose to proteins. As such, it is most necessary to focus on those that interact closely with engineered tissue.
One type of biomolecule that is subject to much research and speculation in current tissue engineering is the growth factor.11 Specific growth factors can have a variety of functions from general cell proliferation to the formation of blood cells and vessels.12-14 They can also be responsible for disease, especially the unchecked cell generation of cancer.15 Many of the positive roles have direct applications to tissue engineering. For example, Transforming Growth Factor-beta (TGF-β) regulates normal growth and development in humans.16 One study found that while addition of ligands to engineered tissue could increase cellular adhesion to nearby cells, the addition also decreased the generation of the extracellular matrix, a key structure in functional tissue.17 To remedy this, the researchers then tested the same method with the addition of TGF-β. They saw a significant increase in the generation of the extracellular matrix, improving their engineered tissue’s ability to become functional faster and more effectively. Clearly, a combination of growth factors and other tissue engineering methods can lead to better outcomes for functional tissue engineering.
With the utility of growth factors established, delivery methods become very important. Several methods have been shown as effective, including delivery in a gelatin carrier.18 However, some of the most promising procedures rely on the scaffolding’s properties. One set of studies mimicked the natural release of growth factors through the extracellular matrix by creating a nanofiber scaffold containing growth factors for delayed release.19 The study saw an positive influence on the behavior of cells as a result of the release of growth factor. Other methods vary physical properties of the scaffold such as pore size to trigger immune pathways that release regenerative growth factors, as will be discussed later. The use of biomolecules and specifically growth factors is heavily linked to the choice of scaffolding material and can be critical to the success of an engineered tissue.
Vascularization
Because almost all tissue cannot survive without proper oxygenation, engineered tissue vascularization has been a focus of many researchers in recent years to optimize chances of engineered tissue success.20 For many of the areas of advancement, this process depends on the scaffold.21 The actual requirements for level and complexity of vasculature vary greatly based on the type of tissue; the requirements for blood flow in the highly vascularized lungs are different than those for cortical bone.22,23 Therefore, it is more appropriate for this topic to address the methods which have been developed for creating vascularized tissue rather than the actual designs of specific tissues.
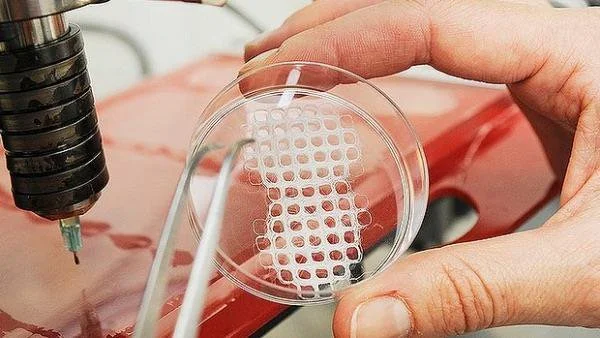
One method that has shown great promise is the use of modified 3D printers to cast vascularized tissue.24 This method uses the relatively new printing technology to create carbohydrate glass networks in the form of the desired vascular network. The network is then coated with a hydrogel scaffold to allow cells to grow. The carbohydrate glass is then dissolved from inside of the hydrogel, leaving an open vasculature in a specific shape. This method has been successful in achieving cell growth in areas of engineered tissue that would normally undergo necrosis. Even more remarkably, the created vasculature showed the ability to branch into a more complex system when coated with endothelial cells.24
However, this method is not always applicable. Many tissue types require scaffolds that are more rigid or have different properties than hydrogels. In this case, researchers have focused on the effect of a material’s porosity on angiogenesis.7,25 Several key factors have been identified for blood vessel growth, including pore size, surface area, and endothelial cell seeding similar to that which was successful in 3D printed hydrogels. Of course, many other methods are currently being researched based on a variety of scaffolds. Improvements on these methods, combined with better research into the interactions of vascularization with biomaterial attachment, show great promise for engineering complex, differentiated tissue.
Mechanical Strength
Research has consistently demonstrated that large-scale cell culture is not limiting to bioengineering. With the introduction of technology like bioreactors or three-dimensional cell culture plates, growing cells of the desired qualities and in the appropriate form continues to become easier for researchers; this in turn allows for a focus on factors beyond simply gathering the proper types of cells.2 This is important because most applications in tissue engineering require more than just the ability to create groupings of cells—the cells must have a certain degree of mechanical strength in order to functionally replace tissue that experiences physical pressure.
The mechanical strength of a tissue is a result of many developmental factors and can be classified in different ways, often based on the type of force applied to the tissue or the amount of force the tissue is able to withstand. Regardless, mechanical strength of a tissue primarily relies on the physical strength of the tissue and its ability for its cells to function under an applied pressure; these are both products of the material and fabrication methods of the scaffolding used. For example, scaffolds in bone tissue engineering are often measured for compressive strength. Studies have found that certain techniques, such as cooking in a vacuum oven, may increase compressive strength.26 One group found that they were able to match the higher end of the possible strength of cancellous (spongy) bone via 3D printing by using specific molecules within the binding layers.27 This simple change resulted in scaffolding that displayed ten times the mechanical strength of scaffolding with traditional materials, a value within the range for natural bone. Additionally, the use of specific binding agents between layers of scaffold resulted in increased cellular attachment, the implications of which will be discussed later.27 These changes result in tissue that is more able to meet the functional requirements and therefore to be easily used as a replacement for bone. Thus, simple changes in materials and methods used can drastically increase the mechanical usability of scaffolds and often have positive effects on other important qualities for certain types of tissue.
Clearly, not all designed tissues require the mechanical strength of bone; contrastingly for contrast, the brain experiences less than one kPa of pressure compared to the for bone’s 106 kPa pressure bones experience.28 Thus, not all scaffolds must support the same amount of pressure, and scaffolds must be made accordingly to accommodate for these structural differences. Additionally, other tissues might experience forces such as tension or torsion based on their locations within the body. This means that mechanical properties must be looked at on a tissue-by-tissue basis in order to determine their corresponding scaffolding structures. But mechanical limitations are only a primary factor in bone, cartilage, and cardiovascular engineered tissue, the latter of which has significantly more complicated mechanical requirements.29
Research in the past few years has investigated increasingly complex aspects of scaffold design and their effects on macroscopic physical properties. For example, it is generally accepted that pore size and related surface area within engineered bone replacements are key to cellular attachment. However, recent advances in scaffold fabrication techniques have allowed researchers to investigate very specific properties of these pores such as their individual geometry. In one recent study, it was found that using an inverse opal geometry--an architecture known for its high strength in materials engineering--for pores led to a doubling of mineralization within a bone engineering scaffold.30 Mineralization is a crucial quality of bone because of its contribution to compressive strength.31 This result is so important because it demonstrates the recent ability of researchers to alter scaffolds on a microscopic level in order to affect macroscopic changes in tissue properties.
Attachment to Nearby Tissue
Even with an ideal design, a tissue’s success as an implant relies on its ability to integrate with the surrounding tissue. For some types of tissue, this is simply a matter of avoiding rejection by the host through an immune response.32 In these cases, it is important to choose materials with a specific consideration for reducing this immune response. Over the past several decades, it has been shown that the key requirement for biocompatibility is the use of materials that are nearly biologically inert and thus do not trigger a negative response from natural tissue.33 This is based on the strategy which focuses on minimizing the immune response of tissue surrounding the implant in order to avoid issues such as inflammation which might be detrimental to the patient undergoing the procedure. This method has been relatively effective for implants ranging from total joint replacements to heart valves.
Avoiding a negative immune response has proven successful for some medical fields. However, more complex solutions involving a guided immune response might be necessary for engineered tissue implants to survive and take on the intended function. This issue of balancing biochemical inertness and tissue survival has led researchers to investigate the possibility of using the host immune response in an advantageous way for the success of the implant.34 This method of intentionally triggering surrounding natural tissue relies on the understanding that immune response is actually essential to tissue repair. While an inert biomaterial may be able to avoid a negative reaction, it will also discourage a positive reaction. Without provoking some sort of response to the new tissue, an implant will remain foreign to bordering tissue; this means that the cells cannot take on important functions, limiting the success of any biomaterial that has more than a mechanical use.
Current studies have focused primarily on modifying surface topography and chemistry to target a positive immune reaction in the cells surrounding the new tissue. One example is the grafting of oligopeptides onto the surface of an implant to stimulate macrophage response. This method ultimately leads to the release of growth factors and greater levels of cellular attachment because of the chemical signals involved in the natural immune response.35 Another study found that the use of a certain pore size in the scaffold material led to faster and more complete healing in an in vivo study using rabbits. Upon further investigation, it was found that the smaller pore size was interacting with macrophages involved in the triggered immune response; this interaction ultimately led more macrophages to differentiate into a regenerative pathway, leading to better and faster healing of the implant with the surrounding tissue.36 Similar studies have investigated the effect of methods such as attaching surface proteins with similarly enlightening results. These and other promising studies have led to an increased awareness of chemical signaling as a method to enhance biomaterial integration with larger implications including faster healing time and greater functionality.
Conclusion
The use of scaffolds for tissue engineering has been the subject of much research because of its potential for extensive utilization in the medical field. Recent advancements have focused on several areas, particularly the use of biomolecules, improved vascularization, increases in mechanical strength, and attachment to existing tissue. Advancements in each of these fields have been closely related to the use of scaffolding. Several biomolecules, especially growth factors, have led to a greater ability for tissue to adapt as an integrated part of the body after implantation. These growth factors rely on efficient means of delivery, notably through inclusion in the scaffold, in order to have an effect on the tissue. The development of new methods and refinement of existing ones has allowed researchers to successfully vascularize tissue on multiple types of scaffolds. Likewise, better methods of strengthening engineered tissue scaffolds before cell growth and implantation have allowed for improved functionality, especially under mechanical forces. Modifications to scaffolding and the addition of special molecules have allowed for increased cellular attachment, improving the efficacy of engineered tissue for implantation. Further advancement in each of these areas could lead to more effective scaffolds and the ability to successfully use engineered tissue for functional implants in medical treatments.